Example for the implemented gass density distributions of the Milky Way¶
The following example shows the use of density distribution for the Milky Way and gives some figures of the distribution.¶
Currently implemented models are * Ferrière: contains HI, HII, H2; in two regions (\(R <3kpc\): arXiv:astro-ph/0702532 ; \(R >3kpc\): ApJ 497:759) * Cordes: contains HII (Nature volume 354, pages 121–124) * Nakanishi: contains HI (arXiv:astro-ph/0304338) and H2 (arXiv:astro-ph/0610769), implemented fit in (arXiv:1607.07886 Appendix C)
In [1]:
from crpropa import *
import numpy as np
import matplotlib.pyplot as plt
In [2]:
# define densities
FER = Ferriere()
NAK = Nakanishi()
COR = Cordes()
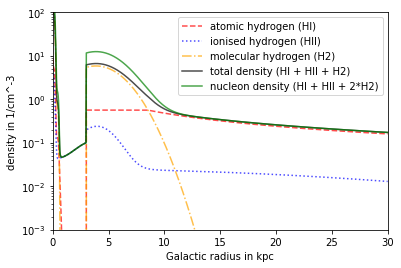
Model Ferrière¶
Contains HI, HII and H2 All types can be deactivated individually, e.g. FER.setisforHX() Dependency in inner Region: x,y,z Dependency in outer Region: R,z
In [3]:
R = np.linspace(0,30*kpc,1000)
phi = np.linspace(0,2*np.pi,360)
n_FER_HI = np.zeros((R.shape[0],phi.shape[0]))
n_FER_HII = np.zeros((R.shape[0],phi.shape[0]))
n_FER_H2 = np.zeros((R.shape[0],phi.shape[0]))
n_FER_tot = np.zeros((R.shape[0],phi.shape[0]))
n_FER_nucl = np.zeros((R.shape[0],phi.shape[0]))
# get densities
pos = Vector3d(0.)
for ir, r in enumerate(R):
for ip, p in enumerate(phi):
pos.x = r*np.cos(p)
pos.y = r*np.sin(p)
n_FER_HI[ir,ip]=FER.getHIDensity(pos)
n_FER_HII[ir,ip]=FER.getHIIDensity(pos)
n_FER_H2[ir,ip]=FER.getH2Density(pos)
n_FER_tot[ir,ip]=FER.getDensity(pos)
n_FER_nucl[ir,ip]=FER.getNucleonDensity(pos)
# plot radial
plt.figure()
plt.plot(R/kpc, n_FER_HI.mean(axis=1)*ccm, linestyle = '--',alpha = .7, color='red', label= 'atomic hydrogen (HI)')
plt.plot(R/kpc, n_FER_HII.mean(axis=1)*ccm, linestyle = ':',alpha = .7, color='blue', label = 'ionised hydrogen (HII)')
plt.plot(R/kpc, n_FER_H2.mean(axis=1)*ccm, linestyle = '-.',alpha = .7, color='orange', label= 'molecular hydrogen (H2)')
plt.plot(R/kpc, n_FER_tot.mean(axis=1)*ccm, color = 'black',alpha = .7, label = 'total density (HI + HII + H2)')
plt.plot(R/kpc, n_FER_nucl.mean(axis=1)*ccm, color ='green',alpha = .7, label = 'nucleon density (HI + HII + 2*H2)')
plt.xlabel('Galactic radius in kpc')
plt.ylabel('density in 1/cm^-3')
plt.yscale('log')
plt.axis([0,30,10**-3,10**2])
plt.legend()
plt.show()
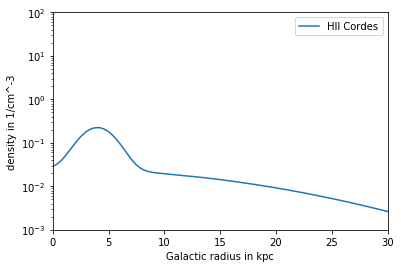
Model Cordes¶
Contains HII component (can not be deactivated) HIIDensity, Density and NucleonDensity are the same Dependency: R,z
In [4]:
n_COR_R= np.zeros(R.shape)
pos = Vector3d(0.)
for ir, r in enumerate(R):
pos.x = r
n_COR_R[ir]= COR.getDensity(pos)
plt.figure()
plt.plot(R/kpc, n_COR_R*ccm, label = 'HII Cordes')
plt.xlabel('Galactic radius in kpc')
plt.ylabel('density in 1/cm^-3')
plt.yscale('log')
plt.axis([0,30,10**-3,10**2])
plt.legend()
plt.show()
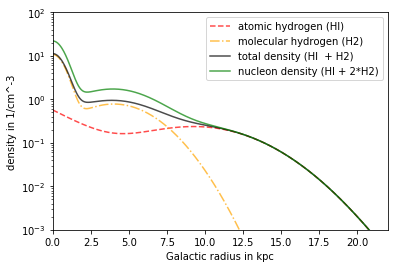
Model Nakanishi¶
Contains HI and H2 Both parts can be deactivated, e.g. setisforHX Dependency: R,z
In [5]:
n_NAK_HI = np.zeros(R.shape)
n_NAK_H2 = np.zeros(R.shape)
n_NAK_tot = np.zeros(R.shape)
n_NAK_nucl= np.zeros(R.shape)
pos = Vector3d(0.)
for ir, r in enumerate(R):
pos.x=r
n_NAK_HI[ir]=NAK.getHIDensity(pos)
n_NAK_H2[ir]=NAK.getH2Density(pos)
n_NAK_tot[ir]=NAK.getDensity(pos)
n_NAK_nucl[ir]=NAK.getNucleonDensity(pos)
# plot radial
plt.figure()
plt.plot(R/kpc, n_NAK_HI*ccm, linestyle = '--',alpha = .7, color='red', label= 'atomic hydrogen (HI)')
plt.plot(R/kpc, n_NAK_H2*ccm, linestyle = '-.',alpha = .7, color='orange', label= 'molecular hydrogen (H2)')
plt.plot(R/kpc, n_NAK_tot*ccm, color = 'black',alpha = .7, label = 'total density (HI + H2)')
plt.plot(R/kpc, n_NAK_nucl*ccm, color ='green',alpha = .7, label = 'nucleon density (HI + 2*H2)')
plt.xlabel('Galactic radius in kpc')
plt.ylabel('density in 1/cm^-3')
plt.yscale('log')
plt.axis([0,22,10**-3,10**2])
plt.legend()
plt.show()
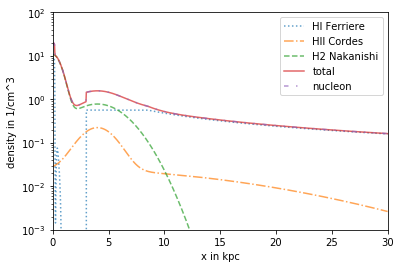
Advanced use of DensityList¶
For example, to combine the HI Component from Ferriere, the HII Component from Cordes and the H2 from Nakanishi
In [6]:
DL = DensityList()
FER.setIsForHII(False)
FER.setIsForH2(False)
DL.addDensity(FER) #only the active HI is added
DL.addDensity(COR) # only the activ HII is added, contains no other typ
NAK.setIsForHI(False)
DL.addDensity(NAK)
# plot types and sum of densities (along x-axis)
n_DL_nucl = np.zeros(R.shape)
n_DL_tot = np.zeros(R.shape)
pos = Vector3d(0.)
for ir, r in enumerate(R):
pos.x = r
n_DL_tot[ir] = DL.getDensity(pos)
n_DL_nucl[ir] = DL.getDensity(pos)
plt.figure()
plt.plot(R/kpc, n_FER_HI[:,0]*ccm, label= 'HI Ferriere', linestyle =':',alpha = .7)
plt.plot(R/kpc, n_COR_R*ccm, label = 'HII Cordes', linestyle ='-.',alpha = .7)
plt.plot(R/kpc, n_NAK_H2*ccm, label='H2 Nakanishi', linestyle = '--', alpha = .7)
plt.plot(R/kpc, n_DL_tot*ccm, label= 'total', linestyle='-',alpha = .7)
plt.plot(R/kpc, n_DL_nucl*ccm, label ='nucleon', linestyle = (0, (3, 5, 1, 5, 1, 5)), alpha = .7)
plt.yscale('log')
plt.xlabel('x in kpc')
plt.ylabel('density in 1/cm^3')
plt.axis([0,30,10**-3,100])
plt.legend()
plt.show()
In [ ]: